Increasing Biopharma ROI with In-House Long-Read Sequencing
September 24, 2024
Raed Hmadi
In the dynamic landscape of biopharmaceutical R&D, next-generation sequencing (NGS) has emerged as an unbiased and highly sensitive technology. It is increasingly being adopted as a multi-attribute method (MAM) to provide information on critical quality attributes (CQAs) such as identity, integrity, and safety. Long-read sequencing has further advanced the field of NGS by enabling the sequencing of long DNA fragments, which was previously problematic due to difficulty sequencing structural variants and complex genomic regions.
Integrating long-read NGS-based assays in-house can significantly streamline biopharma workflows and replace multiple legacy assays with a single quality control assay, which is more efficient and cost-effective. Genedata Selector® is the leading off-the-shelf analysis solution for GMP-compliant NGS-based assays, providing comprehensive, end-to-end support for the entire R&D process. The instrument-agnostic platform automates both short- and long-read NGS data analysis workflows, generating detailed reports for internal evaluation and regulatory submissions.
Advantages of Long-Read Sequencing
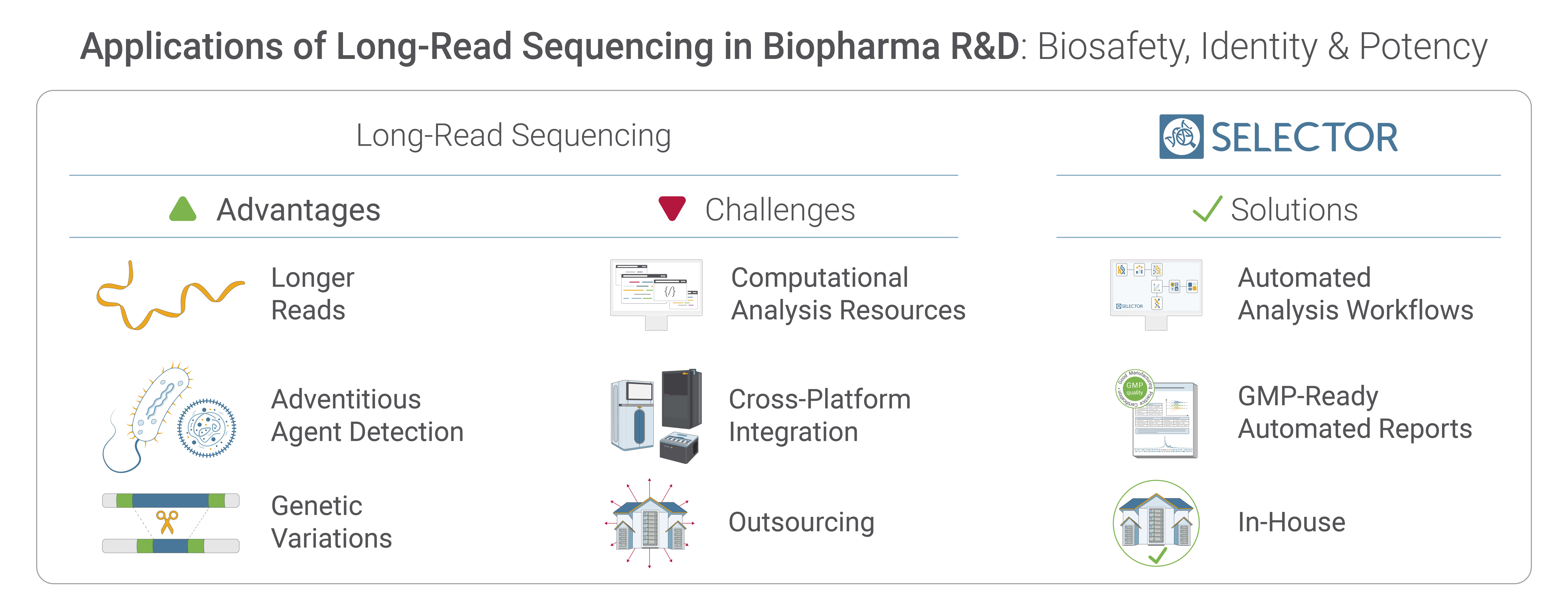
Long-read sequencing, provided by platforms such as Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT), offers numerous benefits over short-read sequencing, extending beyond the ability to accurately sequence longer reads1. Both PacBio and ONT support amplification-free real-time DNA sequencing, eliminating potential bias from PCR amplification. They can also directly identify base modifications such as DNA methylation, bypassing the need for bisulfite conversion. In addition, ONT’s direct RNA sequencing technology circumvents reverse transcription, making RNA isoform identification easier and more efficient2. Given these advantages, long-read sequencing technologies have ushered in a new era of DNA and RNA sequencing.
Applications in Biopharma
Long-read sequencing is being applied in a number of fields, including in biopharma3, agricultural biotechnology4,5, and clinical diagnostics6,7. In biopharma, key applications include clone selection, cell line characterization, genetic stability studies, biosafety testing, and lot release testing8. It is also used to evaluate a wide range of biotherapeutic modalities, including antibodies, vaccines, and cell and gene therapies9.
Safeguarding the integrity of producer cell lines, such as master cell banks, benefits greatly from long-read sequencing. It provides accurate base calling and identifies variants, even in GC-rich repetitive regions, compared to the reference genome. Long-read sequencing is also well suited for verifying over time the genetic identity of engineered cells, such as Chinese hamster ovary (CHO) cells. It captures genetic drift and ensures consistent performance in bioproduction. Additionally, scientists can easily verify gene integration sites and determine gene copy numbers, enabling a comprehensive investigation of cell lines3.
Long-read NGS-based assays are also valuable for developing cell and gene therapies. They detect fusion or truncation events during bioprocess development, ensuring product safety and efficacy. They can also fully sequence plasmids and vectors to verify the orientation of gene inserts, promoters, or variants, regardless of repetitive regions or dimers10. They ensure that correct and error-free adeno-associated viruses (AAVs) are packaged into capsids by thoroughly characterizing AAVs for mutations or deletions, including inverted terminal repeats11.
International regulatory bodies have recently recommended the use of NGS for biosafety testing to replace traditional in vitro and in vivo assays, especially in GMP environments. This shift is driven by the need for more accurate, comprehensive, and efficient testing to ensure the safety and efficacy of therapeutic products. Long-read NGS-based assays offer significant advantages by sequencing native DNA and RNA, spanning entire viral, bacterial, and mycoplasma genomes. This provides a more detailed and complete picture of contaminating genetic material. Long-read sequencing also complements traditional NGS by identifying low abundance contaminants and those with complex genomic structures, improving the detection of known contaminants and the discovery of novel pathogens.
Challenges of Long-Read Sequencing
While the advantages of long-read sequencing are numerous, implementing it in-house presents significant challenges for biopharma organisations. First, the substantial and ultra-rich data generated by long-read sequencing necessitates robust data storage and management solutions. Second, the complexity of long-read data requires specialized bioinformatics tools and expertise, which can be overwhelming in a fast-paced biopharma environment. Automated data analysis workflows with user-friendly interfaces are critical to enable users with diverse bioinformatics backgrounds to quickly learn, perform analyses, and interpret results. Third, extensive computational resources are essential to analyze and process complex data, particularly for genome assembly and variant detection. High-performance computing environments and cloud-based solutions are often necessary, which impacts organizational scalability when processing large datasets. Finally, integrating long-read sequencing data with existing short-read and other omics data is complex but essential for comprehensive quality control testing because it ensures data compatibility and consistency across different sequencing platforms.
Outsourcing long-read NGS-based assays entails a significant investment in time and resources, and comes with IP risks that can cause delays. Analytical R&D teams frequently lack comprehensive access to raw data, hindering further analyses and follow-up investigation, which can lead to problems with unexpected findings or for regulatory submissions. Furthermore, outsourcing challenges include transparency and standardization of assays and workflows, the risk of data quality inconsistencies, and unreliable decision-making. Communication barriers and logistical challenges with external providers further complicate project timelines, data transfer protocols, and quality control measures, all of which leads to potential delays and increased costs.
Genedata Selector Enables In-House Long-Read Sequencing Workflows
Genedata Selector enables biopharma companies to accelerate R&D timelines and increase ROI by streamlining the implementation of in-house long-read NGS-based assays. As the only sequencing platform-agnostic, off-the-shelf analysis solution for GMP-compliant NGS assays, Genedata Selector streamlines the analysis and management of long-read data, enabling the assessment of multiple CQAs, including biosafety, identity, and potency, in a single digital platform. Genedata Selector can integrate all types of sequencing and other omics data and ensures data quality and consistency, making NGS analysis traceable and reproducible.
By leveraging the capabilities of wizard-based Playbooks, Genedata Selector unlocks the potential of long-read sequencing as a MAM approach. Playbooks automate data registration and complex analysis workflows, providing scientists with an intuitive interface and just a few clicks to guide them through all the necessary steps for analyzing and interpreting long-read sequencing results. Additionally, Genedata Selector houses an extensive library of Playbooks covering a wide range of applications and modalities, ensuring comprehensive support for diverse biopharma R&D endeavors. The Playbooks available in the library allow R&D scientists to leverage the power of long-read sequencing for investigating various CQAs including:
- Gene Therapy QC: gene of interest truncation and fusion events, genomic variants, adventitious agent detection, gene expression profiles, vector identity, etc.
- Cell Therapy QC: genomic variants, differential gene expression profiles, single-cell gene expression profiles, plasmid contamination, adventitious agent detection, integration site analysis, copy number variants, etc.
- Master Cell Banks & Cell Line Characterization: gene expression profiles, genomic integrity & stability, product variants, adventitious agent detection, etc.
Genedata Selector comes equipped out-of-the-box with GMP functionalities, providing comprehensive sample history tracking and audit reports. The platform complies with the FDA's 21 CFR Part 11 regulation, ensuring the authenticity, integrity, and confidentiality of records for regulatory submissions. Furthermore, the platform provides scalable data storage and management capabilities, ensuring the seamless handling of large datasets across different sites. This helps to break down data silos and enhances communication and collaboration across teams. Genedata Selector supports enterprise-level scalability for cloud deployment, ensuring that biopharma R&D teams have the computational power needed for large-scale analyses and data interpretation.
Conclusion
Long-read sequencing is transforming the biopharma industry. Deploying long-read NGS-based assays throughout the R&D process marks a pivotal step forward in achieving more streamlined and comprehensive quality control testing. By choosing Genedata Selector, biopharma organizations can cost-effectively harness the potential of long-read sequencing in-house, enabling informed decision-making, enhancing data analysis, and ensuring a higher probability of success. As a single source of truth, Genedata Selector supports biopharma organizations with streamlined regulatory submissions, enhanced data integrity and traceability, and compliance with stringent regulatory standards, all while accelerating R&D timelines and increasing ROI.
References
- Marx, V. (2022). Method of the Year 2022: long-read sequencing. Nature Methods.
- Wang, Y. (2021). Nanopore sequencing technology, bioinformatics and applications. Nature Biotechnology.
- Clappier, C. (2023). Deciphering integration loci of CHO manufacturing cell lines using long read nanopore sequencing. New Biotechnology.
- Pucker, B. (2022). Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions. Quantitative Plant Biology.
- Hamim, I. (2022). How do emerging long-read sequencing technologies function in transforming the plant pathology research landscape? Plant Mol Biol.
- Oehler, J. B. (2023). The application of long-read sequencing in clinical settings. Human Genomics.
- Kobayashi, E. S. (2022). Approaches to long-read sequencing in a clinical setting to improve diagnostic rate. Scientific Reports.
- Logsdon, G. A. (2021). Long-read human genome sequencing and its applications. Nature Reviews Genetics.
- Sripada, S. A. (2024). Advances and opportunities in process analytical technologies for viral vector manufacturing. Biotechnology Advances.
- Hård, J. (2023). Long-read whole-genome analysis of human single cells. Nature Communications.
- Namkung, S. (2022). Direct ITR-to-ITR Nanopore Sequencing of AAV Vector Genomes. Human Gene Therapy.